Inference
CHEESE Inference
The CLI tool supports running CHEESE inference on your custom database. You can just run the command cheese inference and you can check the available options by running cheese inference --help
Usage: -c inference [OPTIONS]
Run CHEESE Inference for an input file.'
╭─ Options ───────────────────────────────────────────────────────────────────────────────────────────────────────────╮
│ * --input_file TEXT The input file in CSV format. Please provide a CSV │
│ file in the following format : 'SMILES,ID │
│ [default: None] │
│ [required] │
│ --dest TEXT Destination folder where to save the results. Will be │
│ inside your source folder │
│ [default: output] │
│ --index_type TEXT Index type : clustered, in_memory, auto │
│ [default: auto] │
│ --gpu_devices TEXT List of GPU devices on which to run computation : e.g │
│ '0,3,2' │
│ [default: 0] │
│ --validate_smiles --no-validate_smiles Whether to validate the SMILES of the input file │
│ [default: no-validate_smiles] │
│ --canonicalize_smiles --no-canonicalize_smiles Whether to canonicalize the SMILES of the input file │
│ [default: no-canonicalize_smiles] │
│ --help Show this message and exit. │
╰─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────╯
The input file should contain lines of molecules in SMILES format and their IDs in the following format : SMILES,ID. Here is an example of an input CSV file.
smiles,id
C[C@H](NC(=O)N1CC2(CCC2)C1c1ccc(F)cc1)C1CC,Z5348285396
CC(NC(=O)N1CC2(CCC2)C1c1ccc(F)cc1)C1CC1,Z5348285396
C[C@@H](NC(=O)N1CC2(CCC2)C1c1ccc(F)cc1)C1CC1,Z5348285396Please note that the index type is defined automatically by default. If the input file exceeds 1GB in size, the script will automatically run the clustered inference, otherwise it will run the in_memory inference.
Example
cheese inference --input_file '/data/mydb.csv' --dest /path/to/my_output --index_type in_memory
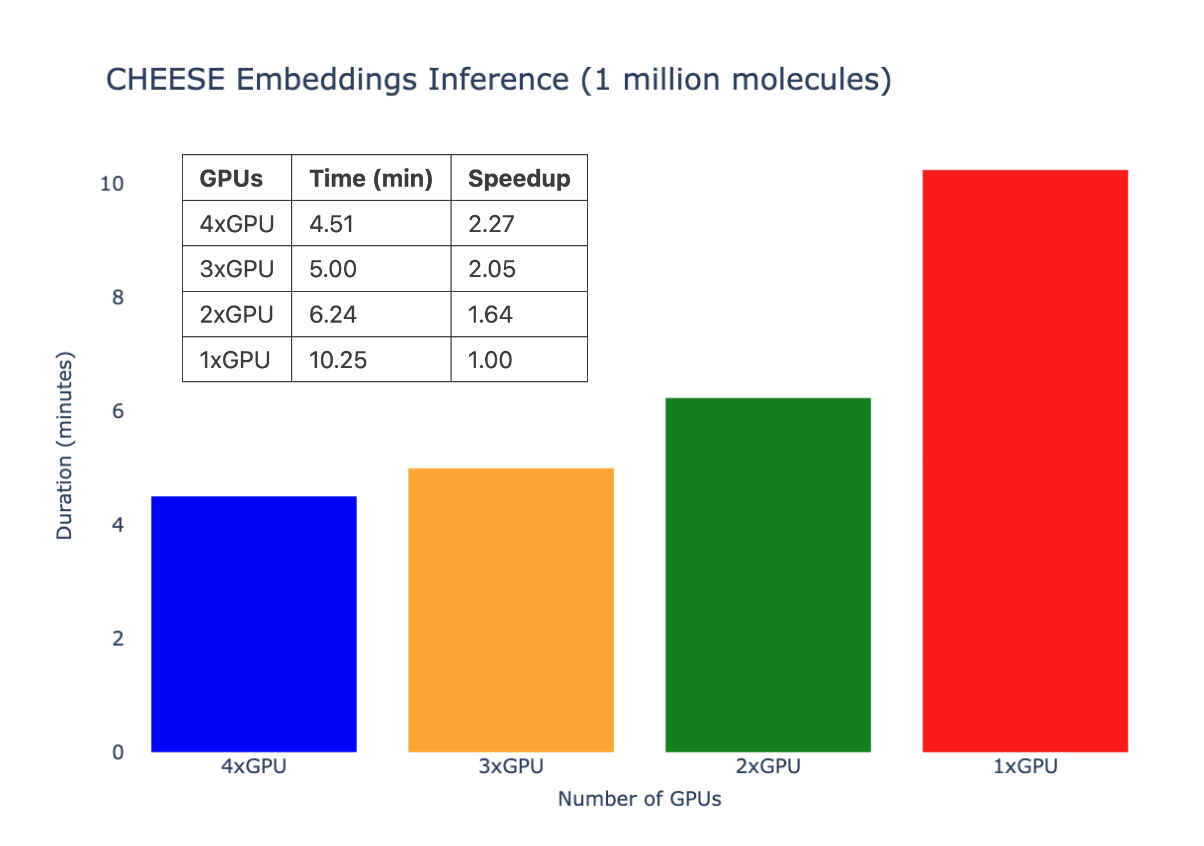
Multiple GPUs inference speed benchmark